Genetic diversity of Capsicum pubescens by functional genomic markers of CYP450
DOI:
https://doi.org/10.29312/remexca.v15i5.3733Keywords:
Capsicum pubescens, genetic variabilityAbstract
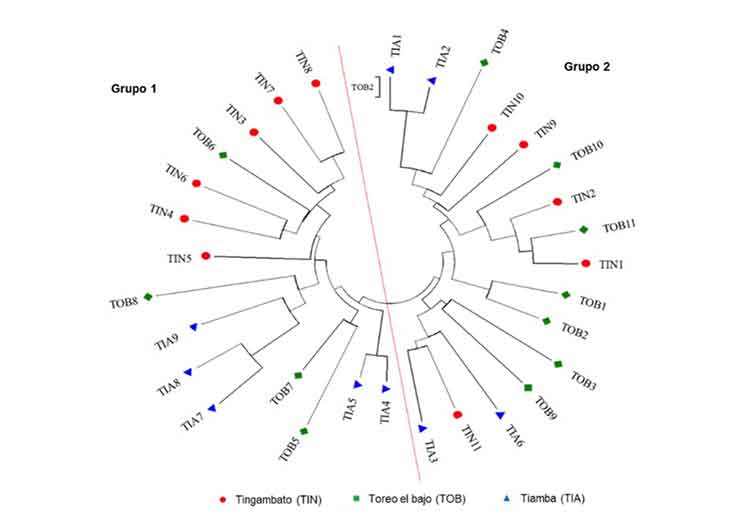
The research work was developed at the Plant Genetic Resources Laboratory of the ‘Presidente Juárez’ Faculty of Agrobiology of the Michoacán University of San Nicolás de Hidalgo, in 2019 and 2022. The aim of this research was to evaluate the usefulness of functional markers derived from CYP450 for genetic diversity studies in Capsicum pubescens. The genetic material consisted of 31 cultivated varieties of C. pubescens from three different localities in the state of Michoacán, Mexico. Genomic DNA was obtained based on the protocol of Huang et al. (2013) and two combinations of CYP450 primers were included in the analysis. The amplification products were separated on 8% acrylamide gels and stained with silver nitrate. A total of 85 loci were detected: the CYP2B6F/CYP2C19R combination detected 34 polymorphic loci, while the CYP2C19F/CYP1A1R combination detected only 27. The diversity analysis of C. pubescens identified 1.54 alleles per locus, 1.33 effective number of alleles per locus, Shannon index of 0.3, a heterozygosity index of 0.2 and 60.39% of polymorphic loci. The results obtained show that markers derived from CYP450 are an efficient and low-cost alternative for studies of genetic diversity in plant species.
Downloads
References
Aguirre, H. E. y Muñoz, O. V. 2015. El chile como alimento. Revista Ciencia. 66(3):16-23. https://www.revistaciencia.amc.edu.mx/images/revista/66-3/PDF/Chile.pdf.
Bak, S.; Beisson, F.; Bishop, G.; Hamberger, B.; Höfer, R.; Paquette, S. and Werck-Reichhart, R. D. 2011. Cytochromes P450. The Arabidopsis Book. 9:2-56. https://doi.org/10.1199/tab.0028.
Bobadilla-Larios, V.; Esparza-Ibarra, E.; Delgadillo-Ruiz, L.; Gallegos-Flores, P. y Ayala-Lujan, J. L. 2017. Variedades de chile (Capsicum annuum L.) identificadas mediante marcadores RAPD. Tropical and Subtropical Agroecosystems. 20(3):465-473. https://www.redalyc.org/pdf/939/93953814014.pdf.
Carvalho, S. I. C.; Ragassi, C. F.; Oliveira, I. B.; Amaral, Z. P. S.; Reifschneider, F. J. B.; Faleiro, F. G. and Buso, G. S. C. 2015. Transferability of microsatellite markers of Capsicum annuum L. to C. frutescens L. and C. chinense Jacq. Genetics and Molecular Research. 14(3):7937-7946. https://doi.org/10.4238/2015.July.17.1.
Castañón-Nájera, G.; Ramírez-Meraz, M.; Ruiz-Salazar, R. y Mayek-Pérez, N. 2011. Aplicación de marcadores AFLP para explorar heterosis en Capsicum spp. Revista Internacional de Botánica Experimental. 80(1):53-58. http://www.revistaphyton.fundromuloraggio.org.ar/vol80/Castanon-Najera-2011.pdf.
Contreras-Toledo, A. R.; López-Sánchez, H.; Santacruz-Varela, A.; Valadez-Moctezuma, E.; Aguilar-Rincón, V. H.; Corona-Torres, T. y López, P. A. 2011. Diversidad genética en México de variedades nativas de chile “poblano” mediante microsatélites. Revista Fitotecnia Mexicana. 34(4):225-232. http://www.scielo.org.mx/scielo.php?pid=S018773802011000400003yscript=sci-arttext.
Dorcas, J. O. F. 2011. Fundamentos de Biología Molecular. Ed. UOC. 1ra. Ed. Barcelona. 244-245 pp.
Escalera-Ordaz, A. K. y Guillén-Andrade, H. 2019. Formas y colores del chile perón. Revista Saber Más. 48(8):42-44. https://www.sabermas.umich.mx/archivo/ articulos/416-numero-48/786-chile-peronvariabilidad-de-formas-y-colores.html.
Gilani, A. S.; Kikuchi, A. and Watanabe, K. N. 2009. Genetic variation within and among fragmented populations of endangered medicinal plant, Withania coagulans (Solanaceae) from Pakistan and its implications for conservation. African Journal of Biotechnology. 8(13):2948-2958. http://www.academicjournals.org/AJB/PDF/ pdf2009/6%20Jul/Gilani%20et%20al.pdf.
Gil-Langarica, H. R. y Mayek-Pérez, N. 2008. Los marcadores moleculares en el mejoramiento genético de la resistencia a enfermedades del frijol (Phaseolus vulgaris L.): aplicaciones y perspectivas. Revista Mexicana de Fitopatología. 26(2):164-176.
Giraldo, H. P. A.; Uribe, S. S. I. y López, R. A. 2011. Análisis de secuencias de ADN mitocondrial (Cytb y ND1) en Lucilia eximia (Diptera: Calliphoridae). Revista Colombiana de Entomología. 37(2):273-278. http://www.scielo.org.co/ pdf/rcen/v37n2/v37n2a20.pdf.
Glasenapp, J. S.; Frieden, B. R. and Cruz, C. D. 2015. Shannon mutual information applied to genetic systems. Quantitative Biology. 1(1):1-20. https://arxiv.org/ftp/arxiv/papers/1512/1512.02324.pdf.
Gonzáles-Mendoza, D. 2009. El complejo enzimático citocromo p450 en las plantas. Revista internacional de contaminación ambiental. 23(4):177-183. http://www.scielo.org.mx/pdf/rica/v23n4/v23n4a3.pdf.
Guzmán, F. A.; Ayala, H. D.; Azurdia, C. A.; Duque, M. C. and De Vicente, M. C. 2005. AFLP assessment of genetic diversity of Capsicum genetic resources in Guatemala: Home gardens as an option for conservation. Crop Science. 45(1):363-370. http://qualquant.org/wp-content/uploads/ethnoecology/2005%20 Guzman363.pdf.
Helliwell, C. A.; Chandler, P. M.; Poole, A.; Dennis, E. S. and Peacock, W. J. 2000. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proceedings of the National Academy of Sciences. USA. 98(4):2065- 2070.
Huang, Q. X.; Wang, X. C.; Kong, H.; Guo, Y. L. and Guo, A. P. 2013. An efficient DNA isolation method for tropical plants. African Journal of Biotechnology. 12(19):2727-2732. https://doi.org/10.5897/AJB12.524.
Inui, H.; Kodama, T.; Ohkawa, Y. and Ohkawa, H. 2000. Herbicide metabolism and cross tolerance in transgenic potato plants co-expressing human CYP1A1, CYP2B6, and CYP2C19. Pesticide Biochemistry and Physiology. 66(2):116-129.
Irwin, D. M.; Kocher, T. D. and Wilson, A. C. 1991. Evolution of the cytochrome b gene of mammals. Journal of Molecular Evolution. 32(2):128-144.
Jaimes-Santoyo, J.; Montesinos-Sampedro, A.; Barbosa-Cobos, R. E.; Moreno-Mutio, S. G.; Rodríguez-Ballesteros, D.; Ramos-Cervantes, T.; Ocharán-Hernández, M. E.; Toscano-Garibay, J. y Beltrán-Ramírez, O. 2014. El citocromo P-450. Revista del Hospital Juárez de México. 81(4):250-256. https://www.medigraphic.com/pdfs/juarez/ju-2014/ju144j.pdf.
Li, G. and Quiros, C. F. 2001. Sequence-related amplified polymorphism (SRAP) a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in brassica. Genética Teórica y Aplicada. 103(2-3):455-461.
Lijun, O. and Xuexiao, Z. 2012. Inter simple sequence repeat analysis of genetic diversity of five cultivated pepper species. African Journal of Biotechnology. 11(4):752-757. https://doi.org/10.5897/AJB10.2551.
López-Espinosa, S. T.; Latournerie-Moreno, L.; Castañón-Nájera, G.; Ruiz-Sánchez, E.; Gómez-Leyva, J. F.; Andueza-Noh, R. H. y Mijangos-Cortés, J. O. 2018. Diversidad genética de chile habanero (Capsicum chinense jacq.) mediante ISSR. Revista Fitotecnia Mexicana. 41(3):227-236. https://doi.org/10.35196/rfm.2018.3.227-236.
Machida-Hirano, R.; Cortés-Cruz, M.; González, B. A. A.; Íñiguez, J. C.; Shirata, K. and Watanabe, K. N. 2015. Isolation and characterization of novel microsatellite markers in chayote [Sechium edule (Jacq.) Sw.]. American Journal of Plant Sciences. 6(13):2033-2041. https://doi.org/10.4236/ajps.2015.613203.
Mahmoud, A. S. 2013. Inter-simple sequence repeat (ISSR) markers in the evaluation of genetic polymorphism of Egyptian Capsicum L. hybrids. African Journal of Biotechnology. 12(7):665-669.
Martínez, C. L. 2007. Reconstrucción de la historia de cambio de los caracteres. Ed. Ecología Molecular. 87-152 pp. https://www.researchgate.net/publication/ 258129643-Ecologia-Molecular.
Panwar, B.; Saini, R. K.; Sharma, N.; Yadav, D. and Kumar, A. 2010. Efficiency of RAPD, SSR and Cytochrome P450 gene-based markers in accessing genetic variability amongst finger millet (Eleusine coracana) accessions. Molecular Biology Reports. https://doi.org/10.1007/s11033-010-0067-5.
Pardey, R. C. y García, D. M. A. 2011. Caracterización molecular de 135 introducciones de Capsicum procedentes del banco de germoplasma de la Universidad Nacional de Colombia sede Palmira. Revista Intropica. 6(1):21-32. https://agris.fao.org/agris-search/search.do?recordID=DJ20220180543.
Peakall, R. and Smouse, P. E. 2012. GenAlex 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics. 28(19):2537-2539. https://doi.org/10.1093/bioinformatics/bts460.
Rentería, A. M. 2007. Breve revisión de los marcadores moleculares. Ed. Ecología Molecular. 541-571. https://hopelchen.tecnm.mx/principal/sylabus/fpdb/ recursos/r119349.PDF.
Ríos, E.; Mejía-Ruiz, H. y Álvarez-Castañeda, S. T. 2009. Marcadores moleculares: una revolución en la Zoología. Revista Ciencia. 60(3):5-13.
Saini, R. K.; Saad, K. R.; Ravishankar, G. A.; Giridhar, P. and Shetty, N. P. 2013. Genetic diversity of commercially grown Moringa oleifera Lam. cultivars from India by RAPD, ISSR and cytochrome P 450-based markers. Plant Systematics and Evolution. 299(7):1205-1213. https://doi.org/10.1007/s00606-013-0789-7.
Sanguinetti, C. J. and Simpson, A. J. 1994. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques. 17(5):914-921.
Shakeel, A. J.; Kikuchi, A.; Ahmad, D. and Watanabe, K. N. 2019. Characterization of the genetic structure of mango ginger (Curcuma amada Roxb.) from Myanmar in farm and genebank collection by the neutral and functional genomic markers. Electronic Journal of Biotechnology. 13(6):1-11. ISSN: 0717-3458.
Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M. and Kumar, S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 28(10):2731-2739. https://doi.org/10.1093/molbev/msr121.
Toledo-Aguilar, R.; López-Sánchez, H.; Santacruz-Varela, A.; Valadez-Moctezuma, E.; López, P. A.; Aguilar-Rincón, V. H.; González-Hernández, V. A. and Vaquera-Huerta, H. 2016. Characterization of genetic diversity of native ‘Ancho’ chili populations of Mexico using microsatellite markers. Chilean Journal of Agricultural Research. 76(1):18-26. https://doi.org/10.4067/S0718-58392016000100003.
Valencia-Quintana, R.; Sánchez-Alarcón, J.; Gómez-Arroyo, S.; Gómez-Olivares, J. L. y Kubiak, S. M. W. 2009. Los citocromos P450 en los 5 reinos de Margulis. Ciencia en la frontera: revista de ciencia y tecnología de la UACJ. 7(1):9-26.
Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; Lee, T.; Van De, Hornes, M. Friters, A.; Pot, J; Paleman, J.; Kuiper, M. and Zabeau, M. 1995. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Research. 23(21):4407-4414. https://doi.org/10.1093/nar/23.21.4407.
Wan, Y.; Watanabe, J. A.; San, S. Y.; Htaik, T.; Win, K.; Yamanaka, S.; Nakamura, I. and Watanabe, K. N. 2005. Assessment of genetic diversity among the major Myanmar banana landraces. Breeding Science. 55(3):365-369. https://www.jstage.jst.go.jp/article/jsbbs/55/3/55-3-365/-pdf.
Whitbred, J. M. and Schuler, M. A. 2000. Molecular characterization of CYP73A9 and CYP82A1 P450 genes involved in plant defense in pea. Plant Physiol. 124(1):47-58.
Winkler, R. G. and Helentjaris, T. 1995. The maize Dwarf3 gene encodes a cytochrome P450- mediated early step in gibberellin biosynthesis. Plant Cell. 7(1):1307-1317.
Xiao-min, Z.; Zheng-hai, Z.; Xiao-zhen, G.; Sheng-li, M.; Xi-xiang, L.; Alain, J. C. P.; Li-Hao, W. and Bao-xi, Z. 2016. Genetic diversity of pepper (Capsicum spp.) germplasm resources in China reflects selection for cultivar types and spatial distribution. Journal of Integrative Agriculture. 15(9):1991-2001.
Yamanaka, S.; Suzuki, E.; Tanaka, M.; Takeda, Y.; Watanabe, J. A. and Watanabe, K. N. 2003. Assessment of cytochrome P450 sequences offers a useful tool for determining genetic diversity in higher plant species. Theoretical and Applied Genetics. 108(1):1-9. https://doi.org/10.1007/s00122-003-1403-0.
Published
How to Cite
Issue
Section
License
Copyright (c) 2024 Revista Mexicana de Ciencias Agrícolas

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
The authors who publish in Revista Mexicana de Ciencias Agrícolas accept the following conditions:
In accordance with copyright laws, Revista Mexicana de Ciencias Agrícolas recognizes and respects the authors’ moral right and ownership of property rights which will be transferred to the journal for dissemination in open access. Invariably, all the authors have to sign a letter of transfer of property rights and of originality of the article to Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias (INIFAP) [National Institute of Forestry, Agricultural and Livestock Research]. The author(s) must pay a fee for the reception of articles before proceeding to editorial review.
All the texts published by Revista Mexicana de Ciencias Agrícolas —with no exception— are distributed under a Creative Commons License Attribution-NonCommercial 4.0 International (CC BY-NC 4.0), which allows third parties to use the publication as long as the work’s authorship and its first publication in this journal are mentioned.
The author(s) can enter into independent and additional contractual agreements for the nonexclusive distribution of the version of the article published in Revista Mexicana de Ciencias Agrícolas (for example include it into an institutional repository or publish it in a book) as long as it is clearly and explicitly indicated that the work was published for the first time in Revista Mexicana de Ciencias Agrícolas.
For all the above, the authors shall send the Letter-transfer of Property Rights for the first publication duly filled in and signed by the author(s). This form must be sent as a PDF file to: revista_atm@yahoo.com.mx; cienciasagricola@inifap.gob.mx; remexca2017@gmail.
This work is licensed under a Creative Commons Attribution-Noncommercial 4.0 International license.


