Yield and stability of soybean genotypes for the tropics of Mexico
DOI:
https://doi.org/10.29312/remexca.v12i8.2267Keywords:
biplot, ideal genotype, relative yieldAbstract
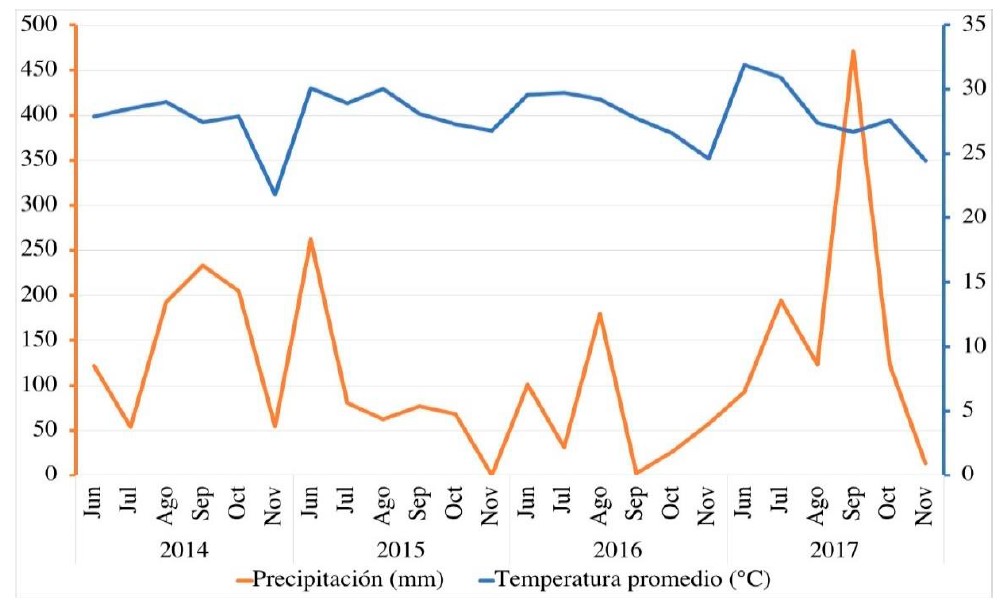
Currently, climate change forces plant breeders to develop genotypes adapted to mega-environments, which guarantees the correct production of the crop. The objective of this study was to determine the potential in grain yield and stability of soybean genotypes. For these purposes, 15 soybean genotypes (seven varieties and eight experimental lines) were evaluated over four years (2014, 2015, 2016 and 2017), in a 5x5 square lattice design with three repetitions. The analysis of variance revealed significant differences in years, genotypes, and in the genotype-by-year interaction. Being the source of variation years, the one that had the greatest impact on yield with 84.3%, followed by the genotype-by-year interaction (10%) and genotypes (5.6%). Likewise, the conditions of 2014 were more conducive for genotypes to have a higher yield. Regarding genotypes, G15, G7, G5 and G2 had the highest yield values throughout the four years. In terms of stability and yield, the relative yield method and the GGE Biplot representation agreed that the genotypes that have these two characteristics are G7 and G2. On the other hand, two mega-environments formed, being genotype G15 the winner with respect to its performance in the first mega-environment, where the years 2015, 2016 and 2017 were included, in the same way, genotype G5 was the winner in the second mega-environment constituted by the year 2014. The two methods when complementing each other mostly explained the phenotypic variation in yield.
Downloads
References
Bhartiya, A.; Aditya, J. P.; Kumari, V.; Kishore, N.; Purwar, J. P.; Agrawal, A. and Kant, L. 2017. GGE biplot & AMMI analysis of yield stability in multi-environment trial of soybean [Glycine max (L.) Merrill] genotypes under rainfed condition of north western Himalayan hills. J. Anim. Plant Sci. 27(1):227-238.
Bhartiya, A.; Singh, K.; Aditya, J. P.; Puspendra and Gupta, M. 2014. Residual relative heterosis and heterobeltiosis for different agro morphological traits as selection index in early segregating generation of soybean [Glycine max (L.) Merrill] crosses. Soybean res. 12(1):28-35.
Brar, K. S.; Singh, P.; Mittal, V. P.; Singh, P.; Jakhar, M. L.; Yadav, Y.; Sharma, M. M.; Shekhawat, U. S. and Kumar, C. 2010. GGE biplot analysis for visualization of mean performance and stability for seed yield in taramira at diverse locations in India. J. Oilseed Brassica. 1(2):66-74.
Casanoves, F.; Baldessari, J. and Balzarini, M. 2005. Evaluation of multienvironment trials of peanut cultivars. Crop Sci. 45(1):18-26. doi:10.2135/cropsci2005.0018. DOI: https://doi.org/10.2135/cropsci2005.0018
Dia, M.; Wehner, T. C.; Hassell, R.; Price, D. S.; Boyhan, G. E.; Olson, S.; King, S.; Davis, A. R. and Tolla, G. E. 2016. Genotype environment interaction and stability analysis for watermelon fruit yield in the United States. Crop Sci. 56(4):1645-1661. doi:10.2135/ cropsci2015.10.0625. DOI: https://doi.org/10.2135/cropsci2015.10.0625
Ebdon, J. S. and Gauch, H. G. 2002. Additive main effect and multiplicative interaction analysis of national turfgrass performance trials. Crop Sci. 42(2):497-506. doi:10.2135/cropsci200 2.0489.
Eberhart, S. T. and Russell, W. A. 1966. Stability parameters for comparing varieties. Crop Sci. 6(1):36-40. doi: 10.2135/cropsci196 6.0011183X000600010011x. DOI: https://doi.org/10.2135/cropsci1966.0011183X000600010011x
Farshadfar, E.; Rashidi, M.; Jowkar, M. M. and Zali, H. 2013. GGE Biplot analysis of genotype environment interaction in chickpea genotypes. Eur. J. Exp. Biol. 3(1):417-423.
Finlay, K. W. and Wilkinson, G. N. 1963. The analysis of adaptation in a plant breeding programme. Aust. J. Agric. Res. 14(6):742-754. doi: 10.1071/AR9630742. DOI: https://doi.org/10.1071/AR9630742
Gauch, H. G.; Piepho, H. P. and Annicchiarico, P. 2008. Statistical analysis of yield trials by AMMI and GGE: Further considerations. Crop Sci. 48(3):866-889. doi:10.2135/cropsci2007. 09.0513. DOI: https://doi.org/10.2135/cropsci2007.09.0513
Haider, Z.; Akhter, M.; Mahmood, A. and Khan, R. A. R. 2017. Comparison of GGE biplot and AMMI analysis of multi-environment trial (MET) data to assess adaptability and stability of rice genotypes. African J. Agric. Res. 12(51):3542-3548. doi:10.5897/AJAR2017.12528. DOI: https://doi.org/10.5897/AJAR2017.12528
Imtiaz, M.; Malhotra, R. S.; Singh, M. and Arslan, S. 2013. Identifying high yielding, stable chickpea genotypes for spring sowing: specific adaptation to locations and sowing seasons in the Mediterranean region. Crop Sci. 53(4):1472-1480. doi:10.2135/cropsci2012.10.0589. DOI: https://doi.org/10.2135/cropsci2012.10.0589
Kang, M. S. 1993. Simultaneous selection for yield and stability in crop performance trials: Consequences for growers. Agron. J. 85(3):754-757. DOI: https://doi.org/10.2134/agronj1993.00021962008500030042x
Kang, M. S. and Gorman, D. P. 1989. Genotype × environment interaction in maize. Agron. J. 81(4):662-664. doi: 10.2134/agron j1989.00021962008100040020x. DOI: https://doi.org/10.2134/agronj1989.00021962008100040020x
Kumudini, S. 2010. Soybean growth and development. The soybean: botany, production and uses. (Ed.) by Guriq Singh. Ludhiana, India. 23 p. DOI: https://doi.org/10.1079/9781845936440.0048
López-Castañeda, C. 2011. Variación en rendimiento de grano, biomasa y número de granos en cebada bajo tres condiciones de humedad del suelo. Trop. Subtrop. Agroecosyst. 14(3):907-918.
López, S. E.; Acosta, G. J. A.; Tosquy, V. O. H.; Salinas, P. R. A.; Sánchez, G. B. M.; Rosales, S. R.; González, R. C.; Moreno, G. T.; Villar, S. B. Cortinas; E. H. M. y Zandate, H. R. 2011. Estabilidad de rendimiento en genotipos mesoamericanos de frijol de grano en México. Rev. Mex. Cienc. Agríc. 2(1):29-40.
Lu’quez, J. E.; Aguirreza’bal, L. A. N.; Aguero, M. E. and Pereyra, V. R. 2002. Stability and adaptability of cultivars in non- balanced yield trials: comparison of methods for selecting ‘high oleic’ sunflower hybrids for grain yield and quality. J. Agron. Crop Sci. 188(4):225-234. doi:10.1046/j.1439-037X.2002.00562.x. DOI: https://doi.org/10.1046/j.1439-037X.2002.00562.x
Magari, R. and Kang, M. S. 1993. Genotype selection via a new yield stability statistic in maize yield trials. Euphytica. 70(1-2):105-111. doi: 10.1007/BF00029647. DOI: https://doi.org/10.1007/BF00029647
Maldonado, M. N. 2017. Soya de temporal y riego para el sur de Tamaulipas ciclo P-V. Agenda Técnica Agrícola Tamaulipas. Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias (INIFAP). Ciudad de México. 92-105 pp.
Mustapha, M. and Bakari, H. R. 2014. Statistical evaluation of genotype by environment interactions for grain yield in millet (Penniisetum glaucum (L.) R. Br.). Int. J. Eng. Sci. 3(9):7-16.
Pecina, Q. V.; Maldonado, H. L.; Maldonado, M. N.; Simpson, J.; Martínez, V. O. y Gil, V. K. C. 2005. Diversidad genética en soya del Trópico Húmedo de México determinada con marcadores AFLP. Rev. Fitotec. Mex. 28(1):63-69.
R. 2020. R: A Language and environment for statistical computing. R foundation for statistical computing. Vienna, Austria. Retrieved from http://www.r-project.org/.
Rimieri, P. 2017. La diversidad y la variabilidad genéticas: dos conceptos diferentes asociados al germoplasma y al mejoramiento genético vegetal. BAG. J. Basic Appl. Gen. 28(2):7-13.
SAS. 2014. SAS: Business analytics and business intelligence software. NC, USA: SAS Inst. Retrieved from http://www.sas.com/en-us/home.html.
Shukla, S.; Mishra, B. K.; Mishra, R.; Siddiqui, A.; Pandey, R. and Rastogi, A. 2015. Comparative study for stability and adaptability through different models in developed high thebaine lines of opium poppy (Papaver somniferum L.). Ind. Crops prod. 74:875-886. doi:10.1016/j.indcrop.2015.05.076. DOI: https://doi.org/10.1016/j.indcrop.2015.05.076
Smith, A. B.; Cullis, B. R. and Thompson, R. 2005. The analysis of crop cultivar breeding and evaluation trials: An overview of current mixed model approaches. J. Agric. Sci. 143(06):449-462. doi: 10.1017/S0021859605005587. DOI: https://doi.org/10.1017/S0021859605005587
Vaezi, B.; Pour-Aboughadareh, A.; Mohammadi, R.; Armion, M.; Mehraban, A.; Hossein-Pour, T. and Dorii, M. 2017. GGE biplot and AMMI analysis of barley yield performance in Iran. Cereal res. Commun. 45(3):500-511. doi:10.1556/0806.45.2017.019. DOI: https://doi.org/10.1556/0806.45.2017.019
Yan, W. 2001. GGE biplot a Windows application for graphical analysis of multienvironment trial data and other types of two-way data. Agron. J. 93(5):1111-1118. DOI: https://doi.org/10.2134/agronj2001.9351111x
Yan, W. and Tinker, N. A. 2005. An integrated biplot analysis system for displaying, interpreting and exploring genotype environment interaction. Crop Sci. 45(3):1004-1016. doi:10.2135/cropsci2004.0076. DOI: https://doi.org/10.2135/cropsci2004.0076
Yan, W. and Kang, M. S. 2003. GGE biplot analysis: a graphical tool for breeders, geneticists, and agronomists. CRC Press. Boca Raton, London, New York, Washington, D.C. retrieved from https://content.taylorfrancis.com/books/download?dac=c2009-0-09571-8&isbn=978 0429122729&format=googlepreviewpdf. 263 p.
Yan, W. 2002. Singular-value partitioning in biplot analysis of multienvironment trial data. Agron. J. 94(5):990-996. doi:10.2134/AGRONJ2002.9900. DOI: https://doi.org/10.2134/agronj2002.0990
Yan, W.; Hunt, L. A.; Sheng, Q. and Szlavnics, Z. 2000. Cultivar evaluation and mega-environment investigation based on the GGE biplot. Crop Sci. 40(3):597-605. doi:10.2135/cropsci2000. 403597x. DOI: https://doi.org/10.2135/cropsci2000.403597x
Yan, W.; Kang, M. S.; Ma, B.; Woods, S. and Cornelius, P. L. 2007. GGE biplot vs. AMMI analysis of genotype-by-environment data. Crop sci. 47(2):643-655. doi:10.2135/cropsci2006. 06.0374. DOI: https://doi.org/10.2135/cropsci2006.06.0374
Yan, W. and Tinker, N. A. 2006. Biplot analysis of multi-environment trial data: principles and applications. Can. J. Plant Sci. 86(3):623-645. doi:10.4141/P05-169. DOI: https://doi.org/10.4141/P05-169
Yang, R. C.; Crossa, J.; Cornelius, P. L. and Burgueño, J. 2009. Biplot analysis of genotype environment interaction: proceed with caution. Crop Sci. 49(5):1564-1576. doi:10.2135/cropsci2008.11.0665. DOI: https://doi.org/10.2135/cropsci2008.11.0665
Yau, S. K. and Hamblin, J. 1994. Relative yield as a measure of entry performance in variable environments. Crop Sci. 34(3):813-817. doi:10.2135/cropsci1994.0011183X0034000 30038x. DOI: https://doi.org/10.2135/cropsci1994.0011183X003400030038x
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2021 Revista Mexicana de Ciencias Agrícolas

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
The authors who publish in Revista Mexicana de Ciencias Agrícolas accept the following conditions:
In accordance with copyright laws, Revista Mexicana de Ciencias Agrícolas recognizes and respects the authors’ moral right and ownership of property rights which will be transferred to the journal for dissemination in open access. Invariably, all the authors have to sign a letter of transfer of property rights and of originality of the article to Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias (INIFAP) [National Institute of Forestry, Agricultural and Livestock Research]. The author(s) must pay a fee for the reception of articles before proceeding to editorial review.
All the texts published by Revista Mexicana de Ciencias Agrícolas —with no exception— are distributed under a Creative Commons License Attribution-NonCommercial 4.0 International (CC BY-NC 4.0), which allows third parties to use the publication as long as the work’s authorship and its first publication in this journal are mentioned.
The author(s) can enter into independent and additional contractual agreements for the nonexclusive distribution of the version of the article published in Revista Mexicana de Ciencias Agrícolas (for example include it into an institutional repository or publish it in a book) as long as it is clearly and explicitly indicated that the work was published for the first time in Revista Mexicana de Ciencias Agrícolas.
For all the above, the authors shall send the Letter-transfer of Property Rights for the first publication duly filled in and signed by the author(s). This form must be sent as a PDF file to: revista_atm@yahoo.com.mx; cienciasagricola@inifap.gob.mx; remexca2017@gmail.
This work is licensed under a Creative Commons Attribution-Noncommercial 4.0 International license.


