Estimation of Podosphaera xanthii in cucumber: machine learning techniques with digital images
DOI:
https://doi.org/10.29312/remexca.v16i30.4039Keywords:
algorithm, disease, powdery mildewAbstract
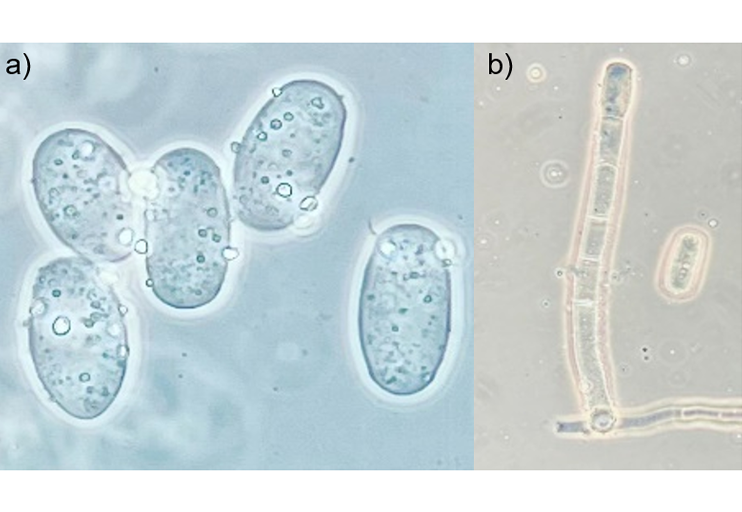
Phytopathogenic fungi pose a considerable threat to cucurbit crops, so early detection and accurate quantification of diseases are essential to reduce production losses. In this study, a methodology was developed to quantitatively estimate the damage caused by Podosphaera xanthii in cucumber leaves, using digital images and machine learning techniques. Convolutional neural networks were used to visually classify the degree of severity into six predefined categories, using sections of leaves with apparent symptoms of the fungus. Additionally, four supervised classification algorithms were trained and compared: K-NN, decision trees, random forests, and neural networks. The model that obtained the best performance was the random forest model, with an accuracy of 90%, whereas K-NN reached the lowest value (79%). These results position the model as a helpful tool for automated disease monitoring in the field, facilitating phytosanitary decision-making. In addition, the methodology provides a solid foundation for researchers interested in designing and implementing automatic plant disease classification systems, providing clear information on the performance of different classification architectures. The algorithm developed in R allows this solution to be adapted and scaled to different cultivation conditions and types of foliar diseases.
Downloads
References
Alaminos, F. A. F. 2023. Árboles de decisión en R con Random Forest. Obts Ciencia Abierta Alicante: limecop. 47-50 pp.
Benali, L.; Notton, G.; Fouilloy, A.; Voyant, C.; Dizene, R.; Boum, H.; Ene, E.; Alia, E.; Ezzouar, B. and Algiers, A. 2019. Solar radiation forecasting using artificial neural network and random forest methods: application to normal beam, horizontal diffuse and global components. Renewable Energy. 132:871-884. https://doi.org/10.1016/j.renene.2018.08.044.
Braun, U. and Cook, A. R. T. 2012. Taxonomic manual of the erysiphales (powdery mildews). Centraalbureau voor Schimmelcultures. 11. CBS Biodiversity: 86-644 pp.
Cipriano, G. R. y González, D. 2022. Identificación molecular de los tipos de compatibilidad en poblaciones de Podosphaera xanthii (Erysiphaceae) infectando cucurbitáceas en Veracruz, México. Acta Botánica Mexicana. 128(129):1-11. https://doi.org/10.21829/abm129.2022.2068.
Cruz, S. H.; Sanchez, M. G.; Rivera, C. J. P. and Avila, G. H. 2020. Identification of phenological stages of sugarcane cultivation using Sentinel-2 images. applications in software engineering proceedings of the 9th. International Conference on Software Process Improvement, CIMPS. 110-116 pp. https://doi.org/10.1109/CIMPS52057.2020.9390095.
Demirovi´c, D. and Stuckey, P. J. 2021. Optimal decision trees for nonlinear metrics. 3733-3744. www.aaai.org.
Fernández, A. 2023. Árboles de decisión en R con Random Forest. Obets Ciencia Abierta. Alicante: limencop. 134 p.
Figueredo, A. A. y Ballesteros, R. J. 2016. Identificación del estado de madurez de las frutas con redes neuronales artificiales, una revisión. 13(1):117-132. https://www.redalyc.org/journal/5600/560062814010/html/.
Flores, P. G.; López, I. F.; Kemp, P. D.; Dörner, J. y Zhang, B. 2016. Modelo de árbol de decisión: una herramienta para el manejo de la pradera. Agro Sur. 44(2):3-10. https://doi.org/10.4206/agrosur.2016.v44n2-02.
Ghawi, R. and Pfeffer, J. 2019. Efficient hyperparameter tuning with grid search for text categorization using knn approach with BM25 similarity. Open Computer Science. 9(1):160-180. https://doi.org/10.1515/COMP-2019-0011/machinereadablecitation/ris.
Guaillazaca, G. C. A. y Hernández, A. V. 2020. Clasificador de productos agrícolas para control de calidad basado en machine learning e industria 4.0. Revista Perspectivas. 2(2):21-28. https://doi.org/10.47187/perspectivas.vol2iss2.pp21-28.2020.
Hassoun, M. H. 1995. Fundamentals of artificial neural networks book. MIT Press. Cambridge, Massachusetts. 417-452 pp. https://kupdf.net/download/48375906-fundamentals-of-artificial-neural-networks-book-1-598b1ef3dc0d601b67300d18-pdf.
Hernández, F. Y.; González, Z. E.; Marrero, T. A. y Dueñas, G. M. J. 2007. Uso de escala para determinar severidad de enfermedades fungosas en híbridos de pepino bajo cultivo protegido. INIFAT. 11(31):49-51.
Kaushik, H.; Khanna, A.; Singh, D.; Kaur, M. and Lee, H. N. 2023. TomFusioNet: a tomato crop analysis framework for mobile applications using the multi-objective optimization based late fusion of deep models and background elimination. Applied Soft Computing. 133:1-24. https://doi.org/10.1016/j.asoc.2022.109898.
Larijani, M. R.; Asli-Ardeh, E. A.; Kozegar, E. and Loni, R. 2019. Evaluation of image processing technique in identifying rice blast disease in field conditions based on KNN algorithm improvement by K-means. Food Science and Nutrition. 7(12):3922-3930. https://doi.org/10.1002/FSN3.1251.
Li, S.; Li, K.; Qiao, Y. and Zhang, L. 2022. A multi-scale cucumber disease detection method in natural scenes based on YOLOv5. Computers and Electronics in Agricultura. 202:1-12. https://doi.org/10.1016/J.COMPAG.2022.107363.
Ma, J.; Du, K.; Zheng, F.; Zhang, L.; Gong, Z. and Sun, Z. 2018. A recognition method for cucumber diseases using leaf symptom images based on deep convolutional neural network. 154:1-7. https://doi.org/10.1016/j.compag.2018.08.048.
Mohamed, Y. F.; Bardin, M. N. P. C. and Pitrat, I. 1995. Causal agents of powdery mildew of cucubits in Sudan. Plant Disease. 79(6):635-636. https://www.apsnet.org/publications/plantdisease/backissues/Documents/1995Articles/PlantDisease79n06-634.
Morejón, G. N.; Coca, M. B. y Martínez, I. D. 2010. Mildiu polvoriento en las cucurbitáceas. Revista de Protección Vegetal. 25(1):44-50. https://revistas.censa.edu.cu/index.php/RPV/article/view/282.
Olivares, B. O.; Vega, A.; Angélica, M.; Calderón, R.; Rey, J. C. and Lobo, D. 2021. Classification of areas affected by banana wilt: an application with machine learning algorithms in Venezuela. https://revistas.up.ac.pa/index.php/REICTORCID.
OTSU, N. A. 1978. Threshold selection method from gray level histogram. IEEE. Transactions on Systems, Man and Cybernetycs. 9(1):62-66.
Pacciorett, P. A.; Kurina, F. G. y Balzarini, M. G. 2020. Muestreo de sitios a escala regional para mapeo digital basado en propiedades de suelo. Ciencia del Suelo. 38(2):310-320. https://www.scielo.org.ar/scielo.php?script=sciarttext&pid=S185020672020000200310&lng=es&tlng=es.
Paymode, A. S. and Malode, V. B. 2022. Transfer learning for multi-crop leaf disease image classification using convolutional neural Network VGG. Artificial Intelligence in Agriculture. 6:23-33. https://doi.org/10.1016/j.aiia.2021.12.002.
Ramos, R. T. V.; Castillo, A. P. J.; Ticona, J. B. y Velasco, B. J. G. 2023. Predicción del éxito del telemarketing bancario mediante el uso de árboles de decisión. Innovación y Software. 4(1):122-137. https://doi.org/10.48168/innosoft.s11.a84.
Rocha, J. F. L.; Reyes, D. Y.; Días, L. E.; Francisco, F. N. y Juárez, C. J. A. 2023. El mildiu polvoriento en calabaza: identificación y manejo bajo las condiciones de Tehuacán, México. Cultivos Tropicales. 44(2): https://cu-id.com/2050/v44n2e09. https://ediciones.inca.edu.cu/index.php/ediciones/article/view/1731.
Sarkar, C.; Gupta, D.; Gupta, U. and Hazarika, B. B. 2023. Leaf disease detection using machine learning and deep learning: review and challenges. Applied Soft Computing. 145:1-61. https://doi.org/10.1016/J.ASOC.2023.110534.
Suganya, D. K.; Srinivasan, P. and Bandhopadhyay, S. 2020. H2K-A robust and optimum approach for detection and classification of groundnut leaf diseases. Computers and Electronics in Agriculture. 178. https://doi.org/10.1016/j.compag.2020.105749.
Sujatha, R.; Chatterjee, J. M.; Jhanjhi, N. Z. and Brohi, S. N. 2021. Performance of deep learning vs machine learning in plant leaf disease detection. Microprocessors and Microsystems. 80:103615. https://doi.org/10.1016/J.MICPRO.2020.103615.
Sun, Z.; Hu, S. Y. and Wen, Y. 2022. Biological control of the cucumber downy mildew pathogen Pseudoperonospora cubensis. Horticulturae. 8:1-15. MDPI. https://doi.org/10.3390/horticulturae8050410.
Torgo L. 2014. Data mining using R: learning with case studies (CRC) Press, Ed. Second. Minneapolis, Minnesota, USA. 87-165 pp. ISBN: 9781439810187.
Velázquez, L. N; Sasaki, Y.; Nakano, K.; Mejía, M. J y Romanchik, K. E. 2011. Detección de cenicilla en rosa usando procesamiento de imágenes por computadora. Revista Chapingo Serie Horticultura. 17(2):151-160. http://sourceforge.net/projects/opencvlibrary.
Wójtowicz, A.; Piekarczyk, J.; Czernecki, B. and Ratajkiewicz, H. 2021. A random forest model for the classification of wheat and rye leaf rust symptoms based on pure spectra at leaf scale. Journal of Photochemistry and Photobiology B. Biology. 223:1-11. https://doi.org/10.1016/j.jphotobiol.2021.112278.
Zhang, S.; Zhang, S.; Zhang, C.; Wang, X. and Shi, Y. 2019. Cucumber leaf disease identification with global pooling dilated convolutional neural network. 162:1-9. https://doi.org/10.1016/j.compag.2019.03.012.
Zhang, Y. and Wallace, B. 2015. A sensitivity analysis and practitioners’ guide to convolutional neural networks for sentence classification. 256-263 pp. http://arxiv.org/abs/1510.03820.
Zhao, Y. and Yang, L. 2023. Distance metric learning based on the class center and nearest neighbor relationship. Neural Networks. 164:631-644. https://doi.org/10.1016/j.neunet.2023.05.004.
Zapata, T. A.; Pérez, L. S. y Mora, F. J. 2014. Método basado en clasificadores k-NN parametrizados con algoritmos genéticos y la estimación de la reactancia para localización de fallas en sistemas de distribución. Revista Facultad de Ingeniería Universidad de Antioquia. 70:220-232. http://www.scielo.org.co/scielo.php?script=sci-arttext&pid=S012062302014000100021&lng=pt&tlng=es.
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Revista Mexicana de Ciencias Agrícolas

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
The authors who publish in Revista Mexicana de Ciencias Agrícolas accept the following conditions:
In accordance with copyright laws, Revista Mexicana de Ciencias Agrícolas recognizes and respects the authors’ moral right and ownership of property rights which will be transferred to the journal for dissemination in open access. Invariably, all the authors have to sign a letter of transfer of property rights and of originality of the article to Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias (INIFAP) [National Institute of Forestry, Agricultural and Livestock Research]. The author(s) must pay a fee for the reception of articles before proceeding to editorial review.
All the texts published by Revista Mexicana de Ciencias Agrícolas —with no exception— are distributed under a Creative Commons License Attribution-NonCommercial 4.0 International (CC BY-NC 4.0), which allows third parties to use the publication as long as the work’s authorship and its first publication in this journal are mentioned.
The author(s) can enter into independent and additional contractual agreements for the nonexclusive distribution of the version of the article published in Revista Mexicana de Ciencias Agrícolas (for example include it into an institutional repository or publish it in a book) as long as it is clearly and explicitly indicated that the work was published for the first time in Revista Mexicana de Ciencias Agrícolas.
For all the above, the authors shall send the Letter-transfer of Property Rights for the first publication duly filled in and signed by the author(s). This form must be sent as a PDF file to: revista_atm@yahoo.com.mx; cienciasagricola@inifap.gob.mx; remexca2017@gmail.
This work is licensed under a Creative Commons Attribution-Noncommercial 4.0 International license.


