Caracterización molecular de Chenopodium berlandieri (Chenopodiaceae) silvestres y cultivados del centro de México
DOI:
https://doi.org/10.29312/remexca.v16i6.3805Palabras clave:
Chenopodium berlandieri subsp. nuttalliae, Chenopodium quinua, marcadores moleculares, SSRResumen
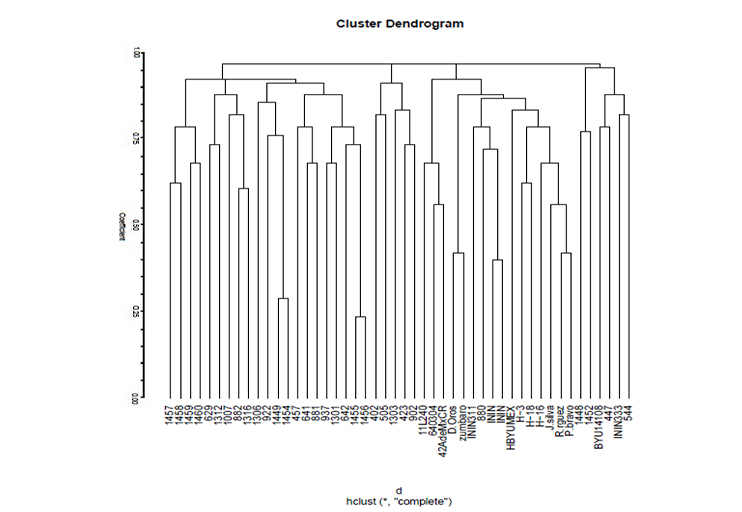
Dentro del género Chenopodium se ubican dos especies de importancia en la alimentación de Mesoamérica y Sudamérica a saber Chenopodium quinoa Willd. (Quinoa) y Chenopodium berlandieri subesp. nuttalliae, cuyos recursos genéticos; no obstante, su gran potencial alimenticio y de adaptabilidad, no han sido caracterizados. Con el objetivo de caracterizar molecularmente germoplasma de Chía roja, Huauzontle (Chenopodium, berlandieri subsp. nuttalliae) y quinoa (Chenopodium quinoa Willd.) se estudiaron molecularmente un total 48 genotipos procedentes de los Bancos de Germoplasma, del Instituto Nacional de Investigaciones Nucleares y del Plant Genetic Resources Laboratory of Brigham Young University. Para determinar la variabilidad genética de Se utilizaron 14 marcadores microsatélites (SSR), específicos para Chenopodium. La afinidad genética se evaluó usando el coeficiente de similitud de Jaccard y el análisis de resultados se realizó mediante el método UPGMA. Los resultados indican que, dentro de los genotipos estudiados de ambas especies se produjeron 175 alelos, que van de 8 (KGA16, QCA88) a 16 (QCA37, QAAT74, QCA57) siendo estos los que más alelos por locus obtuvieron. En el dendrograma se pudo apreciar que a un coeficiente de 0.9 se formaron cuatro grupos principales donde en los grupos 1 y 2 se unen líneas avanzadas de quinua con chía roja, mutantes de chía roja y huauzontle, en los grupos 3 y 4, chía y huauzontle y en el grupo cinco se incluye todo el germoplasma del Plant Genetic Resources Laboratory of BYU, integrado en su mayoría por subespecies de Chenopodium zsachei, boscianum y zinatum. Se concluyó que existe gran afinidad genética entre la quinua, el huauzontle y la chía roja lo que abre posibilidad de realizar cruzas inter e intraespecíficas para el mejoramiento genético de ambas especies.
Descargas
Citas
Allende, C. M. J. 2017. Caracterización morfológica y molecular de accesiones de Quinua (Chenopodium quinoa Willd.) para estimar variabilidad genética. Tesis maestría en mejoramiento genético de plantas. Universidad Nacional Agraria La Molina Escuela de Posgrado. Lima, Perú. 1-90 pp.
Allende, C. L. 2014. Estudio de radiosensibilidad de pseudocereales mediante marcadores moleculares y microscopía electrónica. Tesis Licenciatura, Facultad de ciencias. Universidad Autónoma del Estado de México (UAEM). 17-40 pp.
De-Cruz, T. E.; Xingú, L. A.; García, A. J. M.; Germán, V. I. y Germán, V. G. 2010. Aplicación de técnicas moleculares en el estudio del huauzontle, cultivo prehispánico alternativo para zonas agrícolas. Contacto Nuclear ININ núm. 55. 16-21 pp.
De la Cruz, T. E.; Rubluo, I. A.; Palomino, G. H.; García, A. J. M. and Laguna, C. A. 2007. Characterization of Chenopodium germplasm selection of putative mutants and its cytogenetic study. In: Ochat, S.; Mohan, J. S. Ed. Breeding of neglected and underutilized crops species and herbs. Science Publishers. Enfield, NH, USA. 123-36 pp.
Donaire, T. G. V. 2018. Caracterización molecular de 75 accesiones de quinua (Chenopodium quínoa Willd) del departamento de puno mediante marcadores microsatélites. Tesis Facultad de ciencias. Universidad Nacional Agraria La Molina. Lima, Perú. 122 p.
Eisa, S.; Hussin, S.; Geissler, N. and Koyro, H. W. 2012. Effect of NaCl salinity on water relations, photosynthesis and chemical composition of quinoa (Chenopodium quinoa Willd.) as a potential cash crop halophyte. Australian Journal of Crop Science. 6(2):357-368.
García, A. J. M. 2017. Caracterización molecular de Chenopodium mediante SSR. Informe técnico Científico GB 209/2017. Instituto Nacional de Investigaciones Nucleares, México. 1-3 pp.
Jacobsen, S. E.; Mujica A. and Jensen, C. R. 2003. The resistance of quinoa (Chenopodium quinoa Willd.) to adverse abiotic factors. Food Rev Int. 19(1-2):99-109.
Jacobsen, S. E.; Jensen, C. R. and Liu, F. 2012. Improving crop production in the arid Mediterranean climate. Field Crop Res. 128:34-47. https://doi.org/10.1016/j.fcr.2011.12.001.
Jarvis, D. E.; Kopp, O.; Jellen, E. N.; Marllory, M. C.; Pattee, J.; Bonifacio, F. A.; Coleman, C. E.; Stevens, M. R.; Fairbanks, D. J. and Maughan, P. J. 2008. Simple sequence repeats marker development and genetic mapping in quinoa (Chenopodium quinoa Willd.). J. Genet. 87(1):39-51. https://doi.org/10.1007/s12041-008-0006-6.
Medina, F. J. 2006. Composición nutricional de la quinua. http://xa.yimg.com/kq/groups/21260884/1421089015/name/Composici%C3%B3n+Nutricional+de+la+Quinua.doc.
Morales, A. y Zurita, S. A. 2009. La quinoa como fuente de genes de resistencia a estrés por sequía. Centro Estudios Avanzados en Zonas Áridas, CEAZA. La Serena, Universidad Andrés Bello, Santiago, Chile. 1-8 pp.
Mason, S. L.; Stevens, M. R.; Jellen, E. N.; Bonifacio, F. A.; Fairbanks, D. J.; Coleman C. E.; McCarty, R. R.; Rasmussen, A. G. and Maughan, P. J. 2005. Development and use of microsatellite markers for germplasm characterization in quinoa (Chenopodium quinoa Willd.). Crop Sci. 45(4):1618-1630. https://doi.org/10.2135/cropsci2004.0295.
Maughan, P. J.; Jellen E. R.; Stevens, M. R.; Coleman, C.E.; Ricks, M.; Mason, S. L.; Jarvis, D. E. and Gardunia, B. and Fairbanks, D. J. 2013. Manual. DNA Microprep extraction. Plant genetic resources laboratory of Brigham young university (BYU). Provo, Utah, USA. 1-3 pp.
Maughan, P. J.; Jarvis, D. E.; Cruz-Torres, E.; Jaggi, K. E.; Warner, H. C.; Marcheschi, A. K.; Gomez-Pando, L.; Fuentes, F. ; Mayta-Anko, M. E.; Curti, R.; Rey, E.; Tester, M. and Jellen, E. N. 2024. North American pitseed goosefoot (Chenopodium berlandieri) is a genetic resource to improve Andean quinoa (C. quinoa). Scientific reports. 14:1-13. https://doi.org/10.1038/s41598-024-63106-8.
Nolasco, O. C.; Cruz, W.; Santa-Cruz, C. and Gutiérrez, A. 2013. Evaluation of the DNA polymorphism of six varieties of Chenopodium quinoa Willd, using AFLP. The Biologist. 11(2):277-286.
Ramírez, V. M. L.; Espitia, R. E.; Carballo, C. A.; Zepeda, B. R.; Vaquera, H. H. and Córdova T. L. 2011. Fertilization and plant density in varieties of amaranth (Amaranthus hypochondriacus L.). Revista Mexicana de Ciencias Agrícolas. 2(6):855-866. http://www.redalyc.org/articulo.oa?id=263121473005.
Xingú, L. A. 2010. Caracterización del germoplasma de Huauzontle (Chenopodium berlandieri subsp. nuttalliae) en el Estado de México mediante técnicas moleculares (SSR), Tesis de Maestría, Universidad Autónoma del Estado de México. 9-16 pp.
Xingú-López, A.; Balbuena-Melgarejo, A.; Laguna-Cerda, A. L. G.; Iglesias-Andréu, L. G.; Olivares-Cruz, V. y Cruz-Torres. E. 2018. Caracterización de huauzontle (Chenopodium berlandieri spp. nuttalliae) del Estado de México mediante microsatélites. Ciencia y Tecnol. Agrop. México. 2(6):9-16.
Yasui Y.; Hirakawa, H.; Oikawa, T.; Toyoshima, M.; Matsuzaki, C.; Ueno, M.; Mizuno, N.; Nagatoshi, Y.; Imamura, T.; Miyago, M.; Tanaka, K.; Mise, K.; Tanaka, T.; Mizukoshi, H.; Mori, M. and Fujita, Y. 2016. Draft genome sequence of an inbred line of Chenopodium quinoa, an allotetraploid crop with great environmental adaptability and outstanding nutritional properties. DNA Res. 23(6):535-546. https://doi.org/10.1093/dnares/dsw037.
Descargas
Publicado
Cómo citar
Número
Sección
Licencia
Derechos de autor 2025 Revista Mexicana de Ciencias Agrícolas

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial 4.0.
Los autores(as) que publiquen en Revista Mexicana de Ciencias Agrícolas aceptan las siguientes condiciones:
De acuerdo con la legislación de derechos de autor, Revista Mexicana de Ciencias Agrícolas reconoce y respeta el derecho moral de los autores(as), así como la titularidad del derecho patrimonial, el cual será cedido a la revista para su difusión en acceso abierto.
Los autores(as) deben de pagar una cuota por recepción de artículos antes de pasar por dictamen editorial. En caso de que la colaboración sea aceptada, el autor debe de parar la traducción de su texto al inglés.
Todos los textos publicados por Revista Mexicana de Ciencias Agrícolas -sin excepción- se distribuyen amparados bajo la licencia Creative Commons 4.0 atribución-no comercial (CC BY-NC 4.0 internacional), que permite a terceros utilizar lo publicado siempre que mencionen la autoría del trabajo y a la primera publicación en esta revista.
Los autores/as pueden realizar otros acuerdos contractuales independientes y adicionales para la distribución no exclusiva de la versión del artículo publicado en Revista Mexicana de Ciencias Agrícolas (por ejemplo incluirlo en un repositorio institucional o darlo a conocer en otros medios en papel o electrónicos) siempre que indique clara y explícitamente que el trabajo se publicó por primera vez en Revista Mexicana de Ciencias Agrícolas.
Para todo lo anterior, los autores(as) deben remitir el formato de carta-cesión de la propiedad de los derechos de la primera publicación debidamente requisitado y firmado por los autores(as). Este formato debe ser remitido en archivo PDF al correo: revista_atm@yahoo.com.mx; revistaagricola@inifap.gob.mx.
Esta obra está bajo una licencia de Creative Commons Reconocimiento-No Comercial 4.0 Internacional.


