Identification of proteins in Candidatus Liberibacter asiaticus to develop an immunoenzymatic detection method
DOI:
https://doi.org/10.29312/remexca.v13i8.3355Keywords:
diagnosis, huanglongbing, serologyAbstract
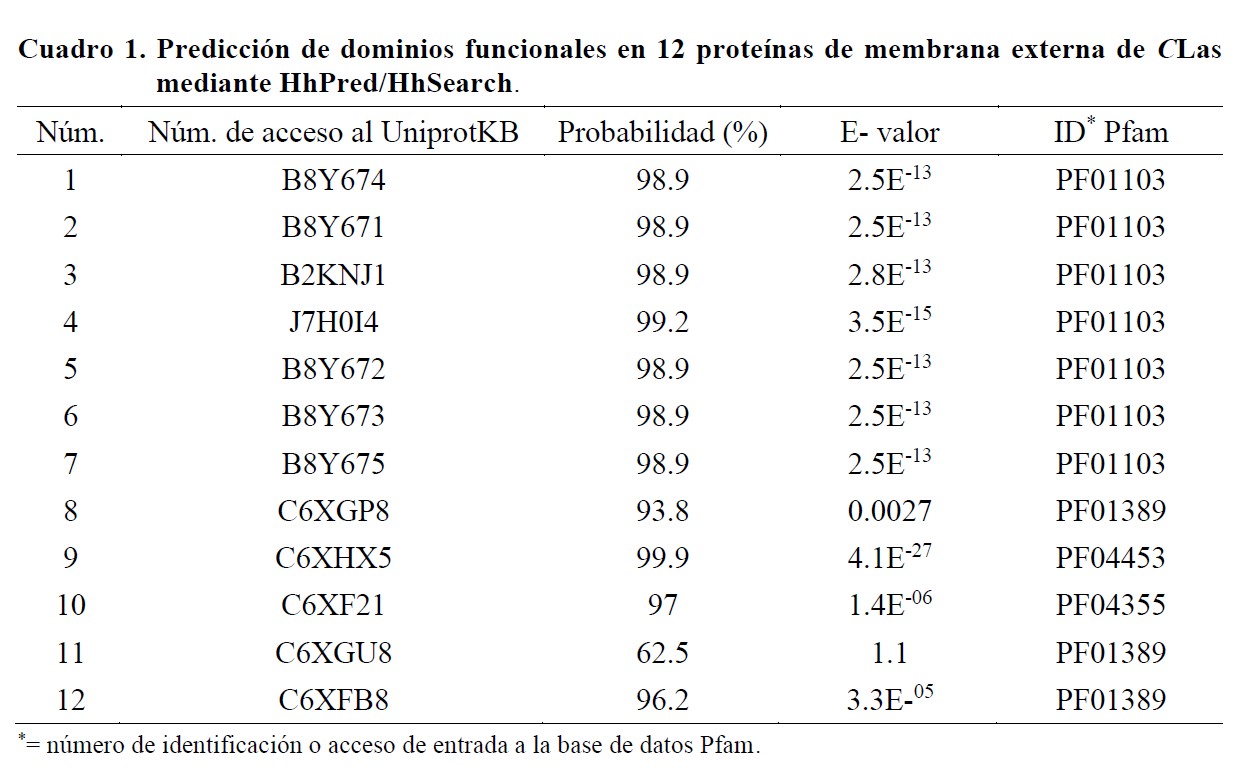
The objective of this work was to identify outer membrane proteins in the genome of Candidatus Liberibacter asiaticus (CLas) with potential for the development and optimization of an immunoenzymatic detection method. The study was conducted during 2019, and the Predict Protein web server, as well as the HhPred/HhSearch and Pfam databases, were used. Fifty-two outer membrane proteins were detected in the complete genome of CLas, of which 11 had not been previously characterized. Predictive analyses performed on the protein B8Y674 generated eight possible epitopes and four of them, experimentally evaluated in B cells, showed percentages of identity between 80 to 90%. CLas was detected by endpoint PCR from DNA extracted from Mexican lime with symptoms of Huanglongbing using primers designed on the sequence of the Omp gene encoding the protein B8Y674 and 95% identity was recorded between the generated sequences and sequences of CLas previously reported. The results obtained allow us to infer that the protein B8Y674 is a potential candidate to be used in the immunoenzymatic detection of CLas.
Downloads
References
Achor, D. S.; Welker, S.; Ben-Mahmoud, S.; Wang, C.; Folimonova. S. Y.; Dutt, M.; Gowda, S. and Levy, A. 2020. Dynamics of Candidatus Liberibacter asiaticus movement and sieve pore plugging in citrus sink cells. Plant Physiol. 182(2):882-891. Doi: 10.1104/pp.19.01391.
Andrade, M. O.; Pang, Z.; Achor, D. S.; Wang, H.; Yao, T.; Singer, B. H. and Wang, N. 2020. The flagella of “Candidatus Liberibacter asiaticus” and its movement in planta. Molecular plant pathology. 21(1):109-123. Doi: 10.1111/mpp.12884.
Bastianel, C.; Garnier-Semancik, M.; Renaudin, J.; Bové, J. M. and Eveillard, S. 2005. Diversity of “Candidatus Liberibacter asiaticus,” based on the omp gene sequence. Appl. Environ. Microbiol. 71(11):6473-6478. Doi: 10.1128/AEM.71.11.6473-6478.2005.
Carrizo, A.; Brihuega, B.; Etchechoury, I.; Arese, A.; Romero, S.; Gioffré, A., Romano, M. F. and Caimi, K. 2009. Identification of immunoreactive antigens of Leptospira interrogans. Rev. Argentina de Microbiol. 41(3):129-133.
Chen, J.; Deng, X.; Sun, X.; Jones, D.; Irey, M. and Civerolo, E. 2010. Guangdong and Florida populations of ‘Candidatus Liberibacter asiaticus’ distinguished by a genomic locus with short tandem repeats. Phytopathology. 100(6):567-572. Doi: 10.1094/PHYTO-100-6-0567.
Conesa, A.; Gotz, S.; García-Gómez. J. M.; Terol, J.; Talón, M. and Robles, M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 21(18):3674-3676.
Ding, F.; Duan, Y.; Paul, C.; Brlansky, R. H. and Hartung, J. S. 2015. Localization and Distribution of “Candidatus Liberibacter asiaticus” in citrus and periwinkle by direct tissue blot immuno assay with an anti ompa polyclonal antibody. Plos One. 10(5):e0123939-10. http://doi.org/ 10.1371/journal.pone.0123939.
Fundora, H. H.; Puig, P. Y.; Chiroles, R. S.; Rodríguez, B. A. M.; Gallardo, D. J. y Milián, S. Y. 2013. Métodos inmunológicos utilizados en la identificación rápida de bacterias y protozoarios en aguas. Rev. Cubana de Hig. Epidemiol. 51(1):84-96.
Lin, H.; Han, C. S.; Liu, B.; Lou, B.; Bai, X.; Deng, C.; Civerolo, E. L. and Gupta, G. 2013. Complete genome sequence of a chinese strain of “Candidatus Liberibacter asiaticus”. Genome Announcements. 1(2):e00184-13. Doi: 10.1128/genomeA.00184-13.
Lu, L.; Cheng, B.; Yao, J.; Peng, A.; Du, D.; Fan, G.; Hu, X.; Zhang, L. and Chen, G. 2013. A new diagnostic system for detection of ‘Candidatus Liberibacter asiaticus’ infection in citrus. Plant Dis. 97(10):1295-1300. http://dx.doi.org/10.1094/ PDIS-11-12-1086-RE.
Mauri, C. and Bosma, A. 2012. Immune regulatory function of b cells. Annual Review of Immunol. 30:221-242. Doi: 10.1146/annurev-immunol-020711-074934.
McCollum, G.; Hilf, M.; Irey, M.; Luo, W. and Gottwald, T. 2016. Susceptibility of sixteen citrus genotypes to ‘Candidatus Liberibacter asiaticus’. Plant Dis. 100(6):1080-1086. http://dx.doi.org/10.1094/PDIS-08-15-0940-RE.
Rodríguez, Q. C. G.; Alanís, M. E. I.; Velázquez, M. J. J. y Almeyda, L. I. H. 2010. Optimización de la técnica de extracción del DNA de plantas de cítricos para el diagnóstico del HLB. En 1er. simposio nacional sobre investigación para el manejo del psílido asiático de los cítricos y el huanglongbing en México. Monterrey, Nuevo León. 22-29 pp.
Rodríguez, Q. C. G.; Almeyda, L. I. H.; Alvarez, O. M. G.; Hernández, G. C. y Mendoza, H. A. 2018. Detección de Candidatus liberibacter asiaticus mediante PCR-punto final, utilizando iniciadores diseñados a partir de los genes omp y clibasia-02425. In: memoria del XXX1 Simposio de avances en Investigación Agrícola, Pecuaria, Forestal, Acuícola, Pesquería, Desarrollo Rural, Transferencia de Tecnología, Biotecnología, Ambiente, Recursos Naturales y Cambio Climático. Veracruz. 1644-1655 pp.
Söding, J.; Biegert, A. and Lupas, A. N. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33(2):244-248. Doi: 10.1093/nar/ gki408.
Tomimura, K.; Miyata, S.; Furuya, N.; Kubota, K.; Okuda, M.; Subandiyah, S.; Hung, T.; Su, H. J. and Iwanami, T. 2009. Evaluation of genetic diversity among ‘Candidatus Liberibacter asiaticus’ isolates collected in southeast Asia. Phytopathology. 99(9):1062-1069. Doi: 10.1094/PHYTO-99-9-1062.
Yuan, Q.; Jordan, R.; Brlansky, R. H.; Minenkova, O. and Hartung, J. 2016. Development of single chain variable fragment (scFv) antibodies against surface proteins of ‘Ca. Liberibacter asiaticus’. J. Microbiol. Methods. 122:1-7. Doi: 10.1016/j.mimet.2015.12.015.
Zherdev, A. V.; Vinogradova, S. V.; Byzova, N. A.; Porotikova, E. V.; Kamionskaya, A. M. and Dzantiev, B. B. 2018. Methods for the diagnosis of grapevine viral infections: a review. Agriculture. 8(12):1-19. Doi: 10.3390/agriculture8120195.
Published
How to Cite
Issue
Section
License
Copyright (c) 2022 Revista Mexicana de Ciencias Agrícolas

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
The authors who publish in Revista Mexicana de Ciencias Agrícolas accept the following conditions:
In accordance with copyright laws, Revista Mexicana de Ciencias Agrícolas recognizes and respects the authors’ moral right and ownership of property rights which will be transferred to the journal for dissemination in open access. Invariably, all the authors have to sign a letter of transfer of property rights and of originality of the article to Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias (INIFAP) [National Institute of Forestry, Agricultural and Livestock Research]. The author(s) must pay a fee for the reception of articles before proceeding to editorial review.
All the texts published by Revista Mexicana de Ciencias Agrícolas —with no exception— are distributed under a Creative Commons License Attribution-NonCommercial 4.0 International (CC BY-NC 4.0), which allows third parties to use the publication as long as the work’s authorship and its first publication in this journal are mentioned.
The author(s) can enter into independent and additional contractual agreements for the nonexclusive distribution of the version of the article published in Revista Mexicana de Ciencias Agrícolas (for example include it into an institutional repository or publish it in a book) as long as it is clearly and explicitly indicated that the work was published for the first time in Revista Mexicana de Ciencias Agrícolas.
For all the above, the authors shall send the Letter-transfer of Property Rights for the first publication duly filled in and signed by the author(s). This form must be sent as a PDF file to: revista_atm@yahoo.com.mx; cienciasagricola@inifap.gob.mx; remexca2017@gmail.
This work is licensed under a Creative Commons Attribution-Noncommercial 4.0 International license.


